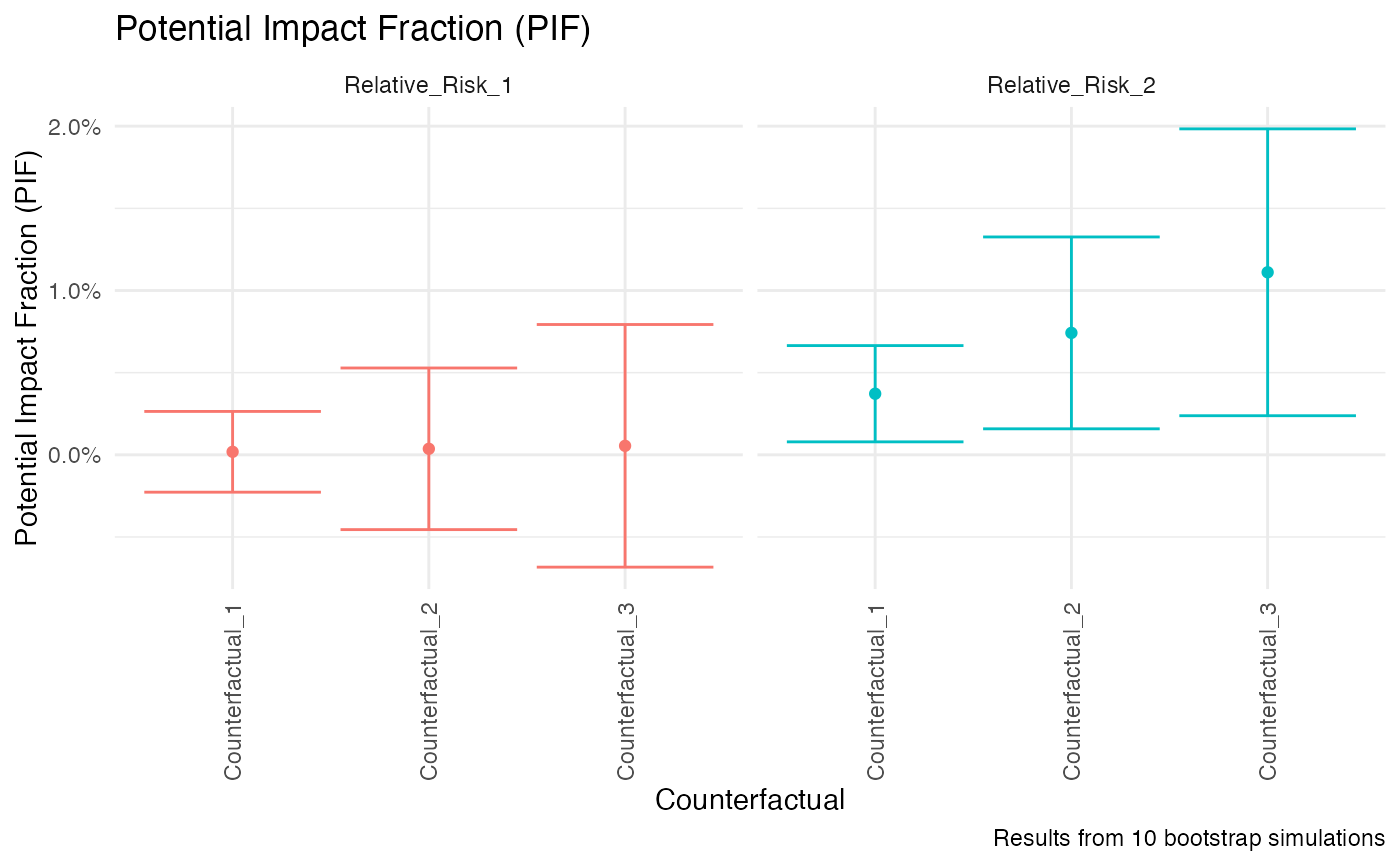
Plot the population attributable fraction or the potential impact fraction
plot.pif_class.RdCreates a ggplot2::ggplot() object containing information on the population
attributable fraction or the potential impact fraction.
Arguments
- x
A
pif_classobject- ...
Additional (currently unused) parameters
- xaxis
Element for the x axis:
counterfactualorrelative_risk- title
The title of the plot
- relative_risks
Vector of relative risk names to plot (set to
NULLto plot all)- counterfactuals
Vector of counterfactual names to plot (set to
NULLto plot all)
Examples
# EXAMPLE 1
# Setup the survey design
data(ensanut)
options(survey.lonely.psu = "adjust")
design <- survey::svydesign(data = ensanut, ids = ~1, weights = ~weight, strata = ~strata)
# Calculate two different relative risks
rr <- list(
function(X, theta) {exp(theta[1] * X[, "systolic_blood_pressure"] / 100)},
function(X, theta) {exp(theta[2] * X[, "systolic_blood_pressure"] / 100 + theta[3]* X[,"age"])}
)
# Calculate three counterfactual scenarios of SBP reduction from 1 to 3 mmhg
cft <- list(
function(X){X[, "systolic_blood_pressure"] <- X[, "systolic_blood_pressure"] - 1; return(X)},
function(X){X[, "systolic_blood_pressure"] <- X[, "systolic_blood_pressure"] - 2; return(X)},
function(X){X[, "systolic_blood_pressure"] <- X[, "systolic_blood_pressure"] - 3; return(X)}
)
my_pif <- pif(design,
theta = log(c(1.05, 1.38, 1.21)), rr, cft,
additional_theta_arguments = c(0.01, 0.03, 0.025),
n_bootstrap_samples = 10,
)
plot(my_pif)
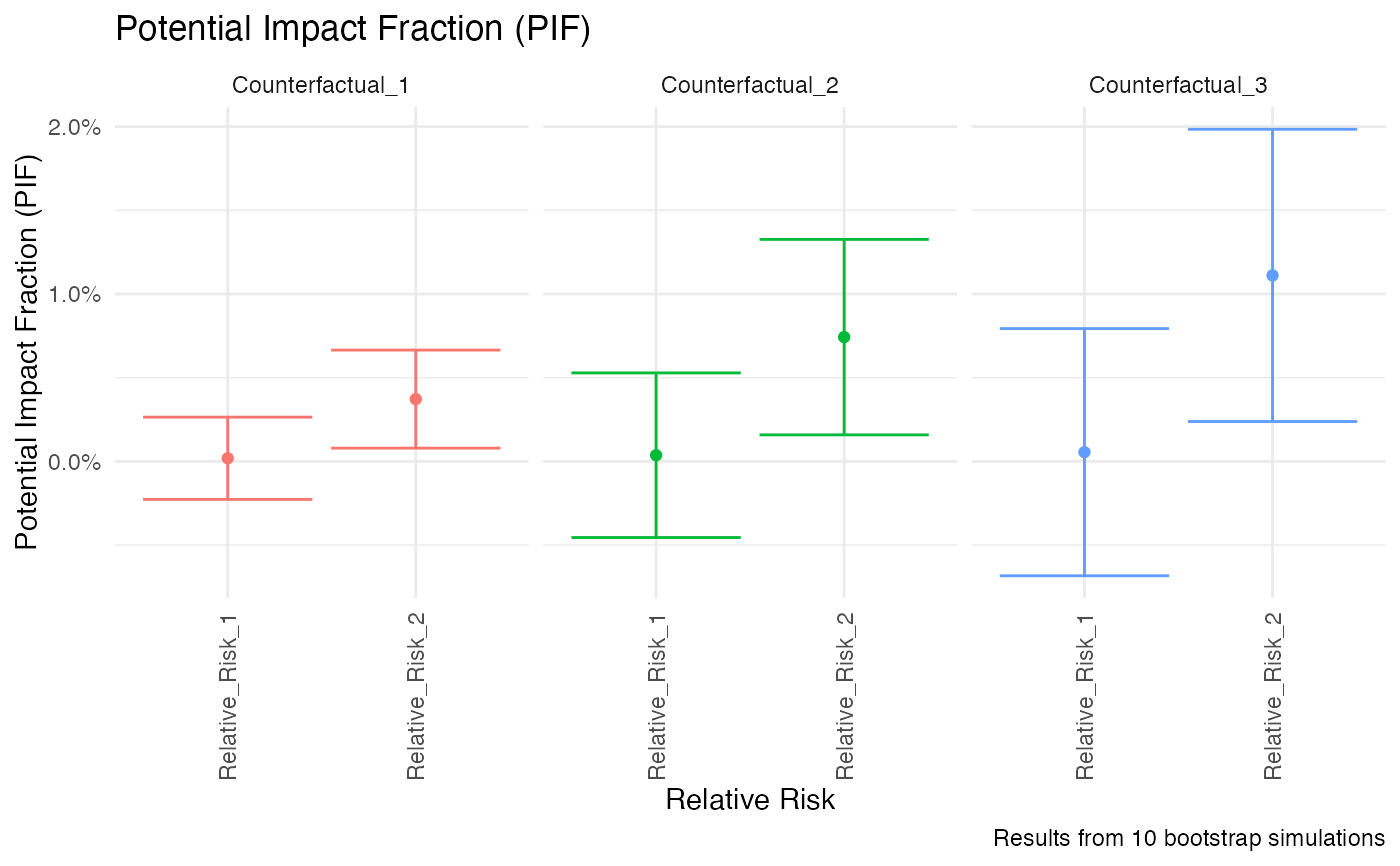 plot(my_pif, xaxis = "counterfactual")
plot(my_pif, xaxis = "counterfactual")